Deep Reinforcement Learning for Protein Folding
The protein structure determines properties and functions of the protein. Therefore, scientistic can develop drugs for this specific unique protein shape in order to cure diseases. Nowadays, Machine Learning methods arise as an alternative to the costly experimentation techniques (cryo-electron microscopy, nuclear magnetic resonance, X-ray crystallography) to help accelerate research.
For the Hands-on Deep Learning for Computer Vision practical course at the Technical University of Munich, we implement the infamous AlphaGo algorithm in C++ to address the protein folding problem with the reinforcement learning approach. The main idea of the algorithm is training a neural network to estimate the policy and the values estimates, where the policy is improved by looking ahead into the future via Monte Carlo Tree Search guided by the value network. The environment (Rosetta 2017.39) and the Monte Carlo Tree Search algorithm were implemented in C++ 17, while the neural network was written in Python (TensorFlow 1.4.1). We rely on the TensorRT library to improve significantly the performance of the neural network inference. As a result, we could speed up the implementation 18.95 times regarding to the python version. Nevertheless, the approach is still unfeasible for large proteins (e.g. L = 332).
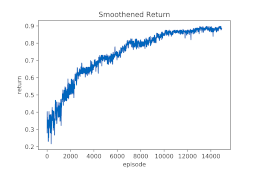
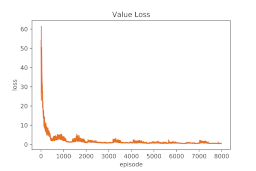
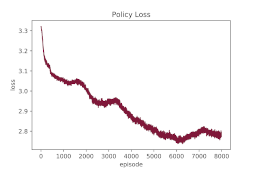
Results overfitting a single protein:
Team Members: Juan Raul Padron Griffe, Matthias Humt, Felix Opolka
Instructors: Vladimir Golkov, Daniel Cremers
DeepMind implemented successfully this project idea in a system called AlphaFold. If you want to know more about cool projects in biomedicine, please contact Vladimir Golkov. Felix Opolka, which was the main author of this project, is currently working on cool publications about neural networks for graph-structured data.